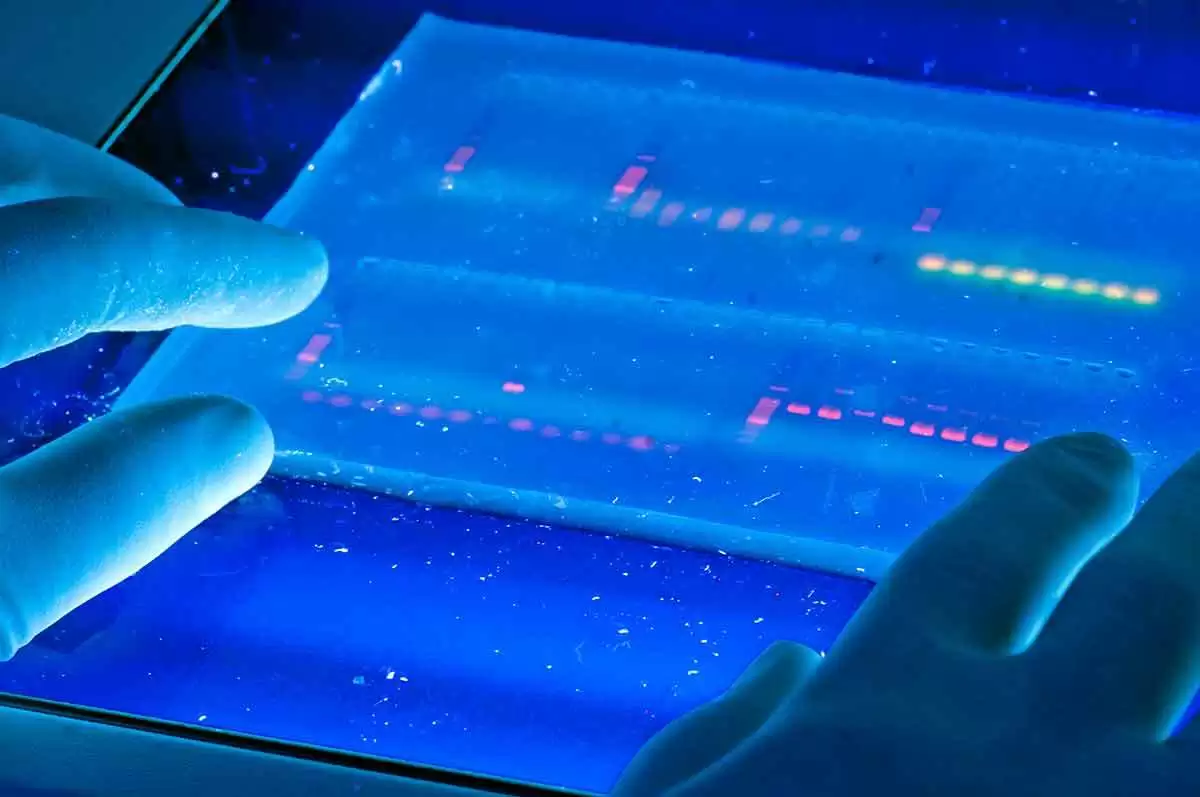
Celiac.com 08/02/2021 - Researchers currently know very little about the causative molecular pathways underlying the development of celiac disease. A team of researchers recently set out to discover new aspects of celiac disease formation and development. To uncover new aspects of disease development, their team used microarrays to measure changes in gene expression of duodenal biopsies.
The team used cDNA microarrays representing 19,200 genes to compare gene expression profiles of duodenal biopsies from 15 celiac disease patients with Marsh III villous atrophy, along with seven control subjects with normal biopsies (Marsh 0). They also looked at the specific effect of gluten by comparing the expression profiles of Marsh III lesions of seven patients exposed to gluten with four patients on a gluten free diet.
Celiac.com Sponsor (A12):
The lesions of Marsh III celiacs versus Marsh 0 control subjects showed that expression levels of 109 genes differed substantially between the two groups.
Many of these genes play roles in proliferation and differentiation pathways, and could be important for proper gut villi development. Changes in these pathways could result in the classic hyperplasia and villous atrophy seen in celiac disease.
The team's comparison patients on a gluten-free diet with those exposed to gluten showed another 120 differentially expressed genes, which could strengthen their observation of increased cell proliferation in the presence of gluten.
The team's findings indicate the role of new candidate genes in the development of celiac disease. Based on their results, they hypothesize that villous atrophy in celiac disease patients arises when cells fail to differentiate properly.
The new candidate genes are involved in pathways not previously implicated in celiac disease development and they may be strong targets for new celiac treatments and therapies. These findings could open new doors for better understanding celiac disease, and lead to new approaches to the study, diagnosis, and treatment of this chronic inflammatory condition.
Read more in Gut. 2004 Jul; 53(7): 944–951.
The research team included Diosdado, M C Wapenaar, L Franke, K J Duran, M J Goerres, M Hadithi, J B A Crusius, J W R Meijer, D J Duggan, C J J Mulder, F C P Holstege, and C Wijmenga. They are variously affiliated with the Complex Genetics Group, Department of Biomedical Genetics, University Medical Centre, Utrecht, the Netherlands; the Complex Genetics Group, and Genomics Laboratory, Department of Biomedical Genetics, University Medical Centre, Utrecht, the Netherlands; the Department of Gastroenterology and Hepatology, Rijnstate Hospital, Arnhem, the Netherlands; the Department of Gastroenterology, Free University Medical Centre, Amsterdam, the Netherlands; the Laboratory of Gastrointestinal Immunogenetics, Free University Medical Centre, Amsterdam, the Netherlands; the Department of Pathology, Rijnstate Hospital, Arnhem, the Netherlands; the National Institute of Arthritis and Musculoskeletal and Skin Diseases, NIH, Bethesda, MD, USA; and the Genomics Laboratory, Department of Physiological Chemistry, University Medical Centre, Utrecht, the Netherlands.






Recommended Comments
Create an account or sign in to comment
You need to be a member in order to leave a comment
Create an account
Sign up for a new account in our community. It's easy!
Register a new accountSign in
Already have an account? Sign in here.
Sign In Now